SCALLOP genetics of the proteome
The SCALLOP consortium (Systematic and Combined AnaLysis of Olink Proteins) is a collaborative framework for discovery and follow-up of genetic associations with proteins on the Olink Proteomics platform. To date, 35 PIs from 28 research institutions have joined the effort, which now comprises summary level data for more than 70,000 patients and controls from 45 cohort studies. SCALLOP welcomes new members.
For more information please contact Anders Mälarstig (SCALLOP project chair).
For the latest news about SCALLOP view this NEWS page,
or read more in our flyer.
Introduction from Anders Mälarstig

Current work
Each SCALLOP member works on human study collections from the general population, clinical trials or patients with certain diseases such as coronary artery disease, rheumatoid arthritis, bipolar disease, heart failure, dementias or metabolic syndrome.
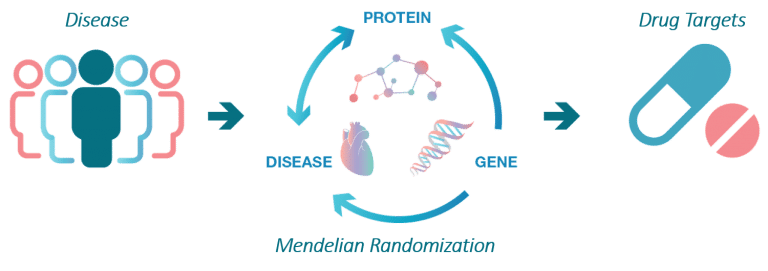
The aim of the SCALLOP consortium is to identify novel molecular connections and protein biomarkers that are causal in diseases.
This work starts with identification of so called protein quantitative trait loci, pQTLs, which are robust connections between a gene variant and the levels of a protein.
There are two types of pQTLs:
- cis-pQTLs are variants that are proximal to the gene encoding the protein under study whereas trans-pQTLs are distal regulation of proteins via an often unknown path.
- Trans-pQTLs can provide unique insights of molecular connections in human biology.
Cis-pQTLs are strong instruments for determining if a protein biomarker for disease is causing disease or elevated or suppressed as a consequence of it. The SCALLOP consortium is currently underway with mapping novel pQTLs for several 100s of proteins in unprecedented sample sizes, something which will yield much deeper insights into the trans-regulation of plasma proteins than what has been possible to date.
Identify causal protein biomarkers
Operations
To be a member of the SCALLOP consortium you have to be the PI of a study collection with Olink proteomics and genome-wide genotyping data. We also expect members to sign up to the Consortium Agreement, which manages conduct and authorships. Download the Consortium Agreement here.
The leadership for subprojects within the SCALLOP consortium rotates and members can take new ideas and suggestions for additional subprojects to the monthly steering committee meetings.
SCALLOP uses a dedicated server for sharing of data. The server is set up under the Danish node of the TRYGGVE server structure. TRYGGVE allows sharing of sensitive data thanks to 2-step authorization procedures and high data security. Thanks to this structure SCALLOP is set up to move to individual-level data should the consortium wish to do so.
Repository browser
| Acronym | Design | Sample size | Olink panels |
| Aristotle | Atrial fibrillation | 1500 | 3 |
| ASAP | Aortic valve surgery | 573 | 5 |
| BAMSE | Childhood asthma | 2000 | 1 |
| Bialystok PLUS | Population based, CVD/CHD | 899 | 5 |
| BioFinder | Dementia | 1550 | 4 |
| CARDIA | Prospective, young adults | 2000 | 2 |
| COSM-C | Population based | 4500 | 3 |
| Dan-NICAD | Stable coronary artery disease | 1650 | 3 |
| DIRECT | Diabetes | 3000 | 5 |
| EpiHealth | Prospective observational | 2500 | 3 |
| Estonian Biobank | Population based | 500 | 4 |
| FENLAND | Population based | 500 | 15 |
| FLEMENGHO | Population based | 3500+ | 1 |
| Framingham | Population based | 520 | 5 |
| HELIC MANOLIS | Population based | 1356 | 5 |
| HELIC POMAK | Population based | 1537 | 5 |
| I AM Frontier | Deep phenotyping, Longitudinal | 240 | 15 |
| IMPROVE | Prospective, metabolic syndrome | 3403 | 1 |
| INTERVAL | Blood donors | 5000 | 4 |
| Kadoorie biobank | Pancreatic cancer | 1400 | 1 |
| KARMA | Incident breast cancer | 1820 | 2 |
| KORA F4 | Population based | 1050 | 1 |
| LBC1936 | Population aged 72 | 750 | 1 |
| LifeLines Deep | Population based | 1200 | 1 |
| Pfizer/ MadCam_ph2 | Inflammatory bowel disease | 200 | 3 |
| MPP-RES | Heart failure | 1000 | 1 |
| NSPHS | Population isolate | 1000 | 6 |
| ORCADES | Population isolate | 1000 | 15 |
| PIVUS | Prospective observational | 933 | 1 |
| POLRED | Population-based, diabetes | 1500 | 5 |
| PRIDE | Dementia | 1500 | 14 |
| PROCARDIS | Coronary artery disease | 900 | 1 |
| PURE | Epidemiological cohort | 8000 | 15 |
| Qmdiab | Diabetes | 320 | 2 |
| RECOMBINE | Rheumatoid arthritis | 800 | 1 |
| RISC | Prospective observational | 1000 | 9 |
| Rotterdam | Population based | 250 | 1 |
| Rotterdam Study-III cohort | Population based | 3500 | 2 |
| SMCC/ SIMPLER | Population based, women | 5000 | 3 |
| STABILITY | Acute coronary syndrome | 3000 | 2 |
| STANLEY | Bipolar disorder, depression | 681 | 4 |
| SWHS/ SMHS/ SCCS | Population based | 300 | 14 |
| ULSAM | Prospective observational | 1000 | 9 |
| VIS | Population isolate | 1000 | 3 |
| WHI | Clinical trial, mixed population, women | 1400 | 6 |
| 1000IBD | Crohn’s disease and ulcerative colitis | 1000 | 1 |
pQTL publications and data from SCALLOP members
Bourgonje, A.R., Hu, S., Spekhorst, L.M., Zhernakova, D.V., Vich Vila, A., Li, Y., ... & Weersma, R.K. (2022). The effect of phenotype and genotype on the plasma proteome in patients with Inflammatory Bowel Disease. Journal of Crohn's and Colitis, 16(3), 414-429. Αrticle link
Macdonald-Dunlop, E., Klarić, L., Folkersen, L., Timmers, P. R., Gustafsson, S., Zhao, J. H., ... & Wilson, J. F. (2021). Mapping genetic determinants of 184 circulating proteins in 26,494 individuals to connect proteins and disease. medRxiv 2021.08.03.21261494. Αrticle link
Lind, L., Gigante, B., Borné, Y., Feldreich, T., Leppert, J., Hedberg, P., ... & Mälarstig, A. (2021). Plasma protein profile of carotid artery atherosclerosis and atherosclerotic outcomes: meta-analyses and mendelian randomization analyses. Arteriosclerosis, Thrombosis, and Vascular Biology, 41(5), 1777-1788. Αrticle link
Smith-Byrne, K., Chen, Y., Kachuri, L., Kapoor, P. M., Guida, F., Zahed, H., ... & Malarstig, A. (2021). IL-18 and Lower Risk for Lung Cancer: Triangulated Evidence from Germline Predictions, Pre-Diagnostic Measurements, and Tumor Expression. medRxiv 2021.03.26.21254400. Αrticle link
Klaric, L., Gisby, J. S., Papadaki, A., Muckian, M. D., Macdonald-Dunlop, E., Zhao, J. H., ... & Peters, J. E. (2021). Mendelian randomisation identifies alternative splicing of the FAS death receptor as a mediator of severe COVID-19. medRxiv 2021.04.01.21254789. Αrticle link
Gisby, J., Clarke, C. L., Medjeral-Thomas, N., Malik, T. H., Papadaki, A., Mortimer, P. M., ... & Peters, J. E. (2020). Longitudinal proteomic profiling of high-risk patients with COVID-19 reveals markers of severity and predictors of fatal disease. medRxiv 2020.11.05.20223289. Αrticle link
Pietzner, M., Wheeler, E., Carrasco-Zanini, J., Raffler, J., Kerrison, N. D., Oerton, E., ... & Langenberg, C. (2020). Genetic architecture of host proteins involved in SARS-CoV-2 infection. Nature communications, 11(1), 1-14. Αrticle link
Folkersen, L., Gustafsson, S., Wang, Q., Hansen, D. H., Hedman, Å. K., Schork, A., ... & Mälarstig, A. (2020). Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nature metabolism, 2(10), 1135-1148. Αrticle link | Read more about the SCALLOP pQTL CVD I study on: SCALLOP news post and KI news.
Suhre, K., McCarthy, M. I., & Schwenk, J. M. (2020). Genetics meets proteomics: perspectives for large population-based studies. Nature Reviews Genetics, 1-19. Αrticle link
Bretherick, A. D., Canela-Xandri, O., Joshi, P. K., Clark, D. W., Rawlik, K., Boutin, T. S., ... & Haley, C. (2020). Linking protein to phenotype with Mendelian Randomization detects 38 proteins with causal roles in human diseases and traits. PLoS genetics, 16(7), e1008785. Αrticle link
Folkersen, L., Gustafsson, S., Wang, Q., Hansen, D. H., Hedman, A. K., Schork, A., ... & Malarstig, A. (2020). Genomic evaluation of circulating proteins for drug target characterisation and precision medicine. BioRxiv 2020.04.03.023804. Article link | Read about the author’s perspective on the SCALLOP pQTL CVD I study on this SCALLOP news post.
Folkersen, L., Fauman, E., Sabater-Lleal, M., Strawbridge, R. J., Frånberg, M., Sennblad, B., ... & Mälarstig, A. (2017). Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS genetics, 13(4), e1006706. Αrticle link
Enroth, S., Johansson, Å., Enroth, S. B., & Gyllensten, U. (2014). Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nature communications, 5(1), 1-11. Αrticle link